| Please, cite us: | Martín-Hernández I, López-Blanco JR, and Chacón P (2025).
Improving the accuracy of Antibody CDR loops structure prediction. (To be submitted)
|
Gallery
Images and media go here.
 Antibody H3 loop (1FGN:HL, H99-106) (8-residues) INPUT: ID= 1FGN:HL, Chain= H, Start= 99, End= 106, Modeling |
 Antigen-Antibody H3 loop (2BDN:AHL, H99-106) (8-residues) INPUT: ID= 2BDN:AHL, Chain= H, Start= 99, End= 106, Modeling |
Help
How to use this application.
Test case: Fetch 6w4w with “6w4w:HL” (or upload the PDB file), choose your CDR loop including Heavy (H) and Light (L) chains, and submit.
Antibody PDB files often include more than just the Heavy (H) and Light (L) chains. These additional
chains may correspond to antigens, multiple Fv fragments, or other molecules.
For example, the PDB entry 1ahw has six chains (A, B, C, D, E and F)
forming two Fv fragments: B/A and E/D (Heavy/Light).
You can find the correspondence between chains and Fv(s) by checking the 1ahw SAbDab
page.
In AbRaCD, you can select exactly which chains to model by simply entering the required Heavy and Light
chain IDs in the corresponding input fields:
First Fv (B/A): Fetch 1ahw with “1ahw:BA” (or upload the PDB file) and set "H chain" to "B" and "L chain" to "A".
Second Fv (E/D): Fetch 1ahw with “1ahw:ED” (or upload the same PDB file) and set
"H chain" to "E" and "L chain" to "D".
Finally, select your desired CDR loop and click Submit. Jobs are queued immediately. Typical runtimes
range from 1–2 minutes for short loops (6–8 residues) up to 20–30 minutes for long loops (≥12 residues).
Any CDR loop can be predicted considering or not the antigen, if present in the PDB file.
For example, the PDB entry 2bdn contains three chains (A, H and L), the Fv
fragment (chains H and L) and the antigen (chain A).
This latter will be considered as long as you include the corresponding chain(s) in the input PDB:
With Antigen: Fetch 2bdn with "2bdn:AHL” (or upload the PDB file).
Without Antigen: Fetch 1ahw with “2bdn:HL” (or upload the PDB file).
In any case, please, set "H chain" to "H" and "L chain" to "L".
Finally, select your desired CDR loop and click Submit. Jobs are queued immediately. Typical runtimes
range from 1–2 minutes for short loops (6–8 residues) up to 20–30 minutes for long loops (≥12 residues).
The improved version of the Random Coordinate Descent (RCD) algorithm is used to generate an ensemble of closed loops (up to 50K in the server) by rotation of the φ and ψ backbone dihedral angles according to detailed neighbor-dependent Ramachandran probability distributions:
Upon loop closure, the loops are evaluated using a fast coarse-grained energy function (ICOSA) and then the best 10% is selected. Finally, these lowest-scoring models are further refined using a detailed energy function (Rosetta) to obtain accurate all-atom predictions.
To perform such loop predictions you only need a minimal basic input: the atomic coordinates of the environment (PDB-file or fetch from PDB) and the boundary residue indices, sequence, and chain of the loop (Step 1). When you press the submit button you will check the job status in the queue tab (Step 3). Once job run is completed you can interactively explore the predicted models in the results tab (Step 4) and/or download them (Step 5). Optionally, advanced users are encouraged to customize RCD+ run by tuning the parameters in the advanced options panel (Step 2).
Please, introduce the following required data and then click on the submit button to
perform the predictions:

(1) Upload your protein atomic structure in PDB format (v3.x) or fetch it by PDB-ID. It is mandatory to follow the PDB format avoiding non standard aminoacids (eg. no CYX, HIE etc.) and atom names. The presence of chain ID is also mandatory. The vast majority of RCD malfunctioning is due to format errors in the PDB. The fetching input format is ID:Chain(s) (In case you don't introduce any chain ID, the first chain ID found in PDB file will be considered). The fetching is very customizable. For example, introduce:
2CMD:A to use the A chain from 2CMD entry,
2BDN:HL to consider the antibody chains H and L of this antigen/antibody complex, or
2BDN:AHL to consider the antigen as well (chain A).
IMPORTANT: This structure will define the loop environment for clash detection and provide the coordinates of anchor aminoacids.
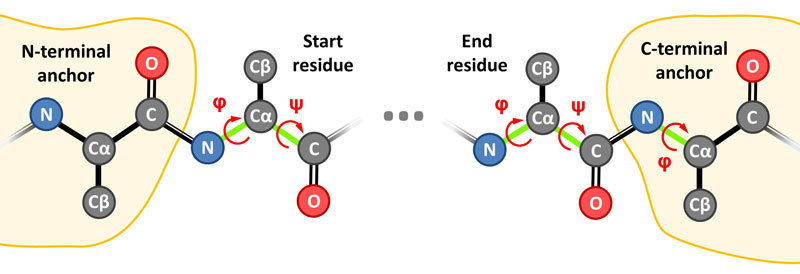
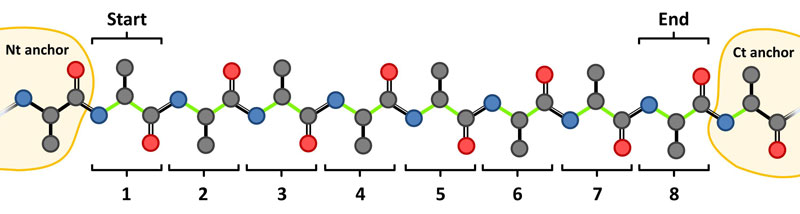
(2) Introduce the chain ID and the Start (first) and End (last) residue indices of the loop. Note that these terminal residues are modeled from scratch and only the N- and C-terminal anchors (indices Start-1 and End+1) and their neighboring residues (indices Start-2 and End+2) are required, e.g. for a 8 residues loop:
Please, use the following indices for the corresponding examples (8 aminoacids long):
A and 270-277 for 2CMD entry or
H and 99-106 for the antigen/antibody case.
(More examples of different loop lengths are provided in the Gallery tab.)
(3) Choose a prediction scenario:
Native if the side-chain conformations of the loop neighborhood are reliable, e.g. building a chrystallographic missing loop or
Modeling to also include the side-chains of the loop neighborhood in the refinement, e.g. predicting loops in homology modeling. As a rule of thumb, Modeling predictions take around three times longer than Native predictions.
(4) Type or paste the sequence of the modeled loop in one letter format (not including anchor aminoacids). This will be the sequence of all modeled loops. Alternatively, leave this field blank to automatically get the sequence from PDB coordinates. Introduce these sequences for the examples:
LGKNGVEE for 2CMD case or
GVFGFFDY for the antigen/antibody complex.
(4) We optionally encourage you to introduce some descriptive job name and an email address for quick results identification and access. It is highly recomended that you use the JAVA interface (JSmol box) in case you have a JAVA virtual machine enabled in the browser since the best 3D visualization performance is only achieved with JAVA. Note that this is not the same as having JavaScript activated. To maximize compatibility with all browsers and devices (Android, iPad or iPhone), please, use HTML5 interface instead (by default).

Inmediately upon job submission, your job will be queued in our server and you will
be redirected to Queue status tab. In this tab all jobs submited to RCD+ Server are
listed. Your jobs are shown in darker colors whereas those submitted by others
appear in lighter colors. You can check server usage and whether any queued job is
running ("r" status, green) or queued ("qw" status, orange). In case any of your
jobs has been queued ("qw" status) it will run as soon as computational resources
become available.

Once a standard-sized job is running ("r" status) it usually takes around 10-15 minutes to complete depending on its size. For example, the modeling of a large loop (12-residues) usually takes around 10 minutes. To avoid server overloading the wall-time of Native and Modeling jobs has been set to 2 and 4 hours, respectively.

As soon as your jobs finish they will move to the list of "Your finished jobs" for further access and a direct link will appear to redirect you to the Results of the last finished job.

In case you detect any problem in any of your submitted jobs they can be easily deleted by clicking the corresponding red cross. A "dr" status (black) will evidence that it is being deleted from queue. Note that anyone but you can delete your jobs.
Please, do not close the browser unless you have either kept track of the job ID or provided an email address, otherwise you will not be able to access your results when the web browser is closed.
Use mouse
controls to interactively explore the generated loops in JSmol and customize
their molecular representation and colors. For example, just drag for rotating, hold
Shift key + double click for translating, or click in the palette to change the
color of the selected loop(s). The user can choose between three different
interfaces: Java, HTML5 and WebGL. The HTML5-javascript interface is activated by
default. To activate the JAVA, first you must enable it in your web browser (details) and add
frodock.chaconlab.org to the exception
sites in the Java panel (details).

You can also check the structural qualitiy of the generated loops by deploying the More information section.

Only if you included the native loop in the PDB (just for benchmarking purposes) the result error in RMSD is evaluated and plotted versus the loop energy for all loop models.

Finally, all computed results are freely avaible for download:

- All refined loops: All-atom refined models as a single Multi-PDB file.
- All closed loops: All closed loop models produced by RCD+ (before any Rosetta refinement) at selected Coarse-Graining level (a single Multi-PDB file too).
- Dunbrack plots: Neighbor-dependent Ramachandran maps, dihedral angles, and images are provided as plain text or JPG images.
- Energy evaluation: Refinement summary table for all refined loops (plain text file) with the loop indices (#loop), the all-atom Rosetta energy before any refinement for the full protein (E_full) and the loop (E_loop), the all-atom Rosetta energy after refinement (E_full2), the RMSDs before (before E_full2 column) and after (after E_full2 column) refinement considering N,Cα,C,O atoms (R_rcd and R_BBO), N,Cα,C (R_BB), all heavy atoms (R_HA), all side-chain heavy atoms (R_SC), and all atoms including hydrogens (R_ALL).
- Summary results: RCD+'s output summary.
- Log-file: Plain text log file with the start/end time for each of the protocol steps.
- All computed files: A tar+gzip file containing all job data.